lattices package
The Lattice class
- class lattices.Lattice(*, name=None, lattice_constants=None, nodal_positions=None, edge_adjacency=None, edge_coordinates=None, fundamental_edge_adjacency=None, fundamental_tesselation_vecs=None, **kwargs)[source]
Bases:
object- property UC_volume: float
Calculate unit cell volume from internally-stored crystal data
Given internally-stored crystal data \(a,b,c,{\alpha},{\beta},{\gamma}\), calculate unit cell volume as
\[abc \sqrt{1 - \cos({\alpha})^2 - \cos(\beta)^2 - \cos(\gamma)^2 + 2\cos(\alpha)\cos(\beta)\cos(\gamma)}\]For more details see the supporting information of PNAS paper at https://www.pnas.org/doi/10.1073/pnas.2003504118
- __init__(*, name=None, lattice_constants=None, nodal_positions=None, edge_adjacency=None, edge_coordinates=None, fundamental_edge_adjacency=None, fundamental_tesselation_vecs=None, **kwargs) None[source]
Construct lattice unit cell.
Takes in keyword-only arguments.
Examples
- Three ways of initialisation:
by specifying node coordinates and edge adjacency
>>> import numpy as np >>> nodes = np.array([[0,0,0],[1,0,0],[0.5,1,0],[0.5,0.5,1]]) >>> edges = np.array([[0,1],[1,2],[0,2],[0,3],[1,3],[2,3]]) >>> lat = Lattice(nodal_positions=nodes, edge_adjacency=edges, >>> name='pyramid') >>> lat {'name': 'pyramid', 'num_nodes': 4, 'num_edges': 6}
by specifying edge coordinates
>>> edge_coords = np.array([ >>> [0,0,0,1,0,0], >>> [1,0,0,0.5,1,0], >>> [0.5,1,0,0,0,0], >>> [0,0,0,0.5,0.5,1], >>> [1,0,0,0.5,0.5,1], >>> [0.5,1,0,0.5,0.5,1] >>> ]) >>> lat = Lattice(edge_coordinates=edge_coords, name='pyramid') >>> lat {'name': 'pyramid', 'num_nodes': 4, 'num_edges': 6}
by specifying fundamental representation
>>> # simple cubic in fundamental representation >>> nodes = [[0.5,0.5,0.5]] >>> fundamental_edge_adjacency = [[0,0],[0,0],[0,0]] >>> tess_vecs = [[0,0,0,1,0,0],[0,0,0,0,1,0],[0,0,0,0,0,1]] >>> lat = Lattice( >>> nodal_positions=nodes, >>> fundamental_edge_adjacency=fundamental_edge_adjacency, >>> fundamental_tesselation_vecs=tess_vecs >>> ) >>> lat.crop_unit_cell() >>> lat {'num_nodes': 7, 'num_edges': 6}
If both edge adjacency and fundamental edge adjacency are specified, simple checks are performed that the fundamental representation is consistent with the edge adjacency. In this case, lattice must be specified in a valid windowed configuration.
Lattice can also be initialised by directly unpacking the catalogue dictionary
>>> from data import Lattice, Catalogue >>> cat = Catalogue.from_file('Unit_Cell_Catalog.txt', 1) >>> lat = Lattice(**cat.get_unit_cell(cat.names[0])) >>> lat {'name': 'cub_Z06.0_E1', 'num_nodes': 8, 'num_edges': 12}
- apply_nodal_imperfections(dr_mag: float, kind: str) Lattice[source]
Displace inner nodes using the fundamental representation.
- Parameters:
dr_mag (float) – magnitude of perturbation
kind (str) – ‘sphere_surf’ or ‘sphere_solid’ or ‘gaussian’
- Returns:
modified lattice with nodal imperfections
- Return type:
Examples
>>> nodes = [[0.3,0.3,0.5],[0.7,0.3,0.5],[0.3,0.7,0.5],[0.7,0.7,0.5]] >>> fundamental_edge_adjacency=[[0,1],[1,3],[2,3],[0,2],[2,0],[3,1],[1,0],[3,2]] >>> tess_vecs = [[0,0,0,0,0,0],[0,0,0,0,0,0],[0,0,0,0,0,0],[0,0,0,0,0,0], >>> [0,0,0,0,1,0],[0,0,0,0,1,0],[0,0,0,1,0,0],[0,0,0,1,0,0]] >>> lat = Lattice( >>> nodal_positions=nodes, >>> fundamental_edge_adjacency=fundamental_edge_adjacency, >>> fundamental_tesselation_vecs=tess_vecs >>> ) >>> lat.crop_unit_cell() >>> lat_imp = lat.apply_nodal_imperfections(0.1, 'sphere_surf')
(
Source code,png,hires.png,pdf)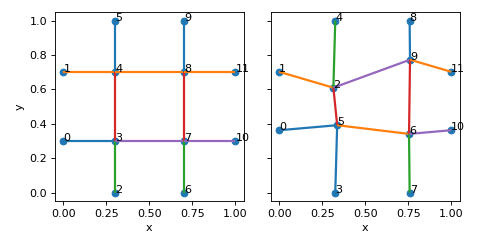
- calculate_edge_lengths(edge_representation: str = 'cropped', coords: str = 'reduced') ndarray[tuple[int, ...], dtype[_ScalarType_co]][source]
Calculate edge lengths in a given representation
- Parameters:
edge_representation (str, optional) – Use edges cropped to fit within unit cell window (‘cropped’) or fundamental edge representation (‘fundamental’). Defaults to ‘cropped’.
coords (str, optional) – Use nodal coordinates in ‘reduced’ or ‘transformed’ coordinate system. Defaults to ‘reduced’.
- Raises:
ValueError – if input parameters are not from the allowed set
- Returns:
edge lengths of shape (num_edges,) if edge_representation`is ‘cropped’ or (num_fundamental_edges,) if `edge_representation is ‘fundamental’.
- Return type:
np.ndarray
- calculate_edge_radius(rel_dens: float, coords: str = 'transformed') float[source]
Calculate uniform edge radii according to relative density.
\[r = \sqrt{\frac{\bar{\rho} \ V}{\pi L}}\]where \(\bar{\rho}\) is the relative density, \(V\) is volume of the unit cell, and \(L\) is sum of the edge lengths.
- Parameters:
rel_dens (float) – target relative density
coords (str, optional) – Use ‘transformed’ or ‘reduced’ coordinates. Defaults to ‘transformed’.
- Returns:
edge radius that gives the target relative density
- Return type:
float
- calculate_fundamental_representation() None[source]
Calculate the fundamental representation of lattice.
The fundamental representation is based on inner nodes and tesselation vectors.
Examples
>>> nodes = [[0,0.5,0.5],[1,0.5,0.5],[0.5,0,0.5],[0.5,1,0.5],[0.5,0.5,0.5]] >>> edges = [[0,4],[1,4],[2,4],[3,4]] >>> lat = Lattice(nodal_positions=nodes, edge_adjacency=edges) >>> lat.calculate_fundamental_representation() >>> lat.node_types {'corner_nodes': set(), 'edge_nodes': set(), 'face_nodes': {0, 1, 2, 3}, 'inner_nodes': {4}} >>> lat.fundamental_edge_adjacency [[4 4] [4 4]] >>> lat.fundamental_tesselation_vecs [[0. 0. 0. 1. 0. 0.] [0. 0. 0. 0. 1. 0.]] >>> lat.num_fundamental_edges 2
(
Source code,png,hires.png,pdf)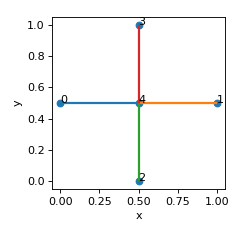
- We see that this lattice has one inner node (4), and two fundamental edges:
4 –> 4 + [1 0 0] (horizontal edge connecting node 4 to its tesselation by vector (1,0,0))
4 –> 4 + [0 1 0] (vertical edge connecting node 4 to its tesselation by vector (0,1,0))
- calculate_nodal_connectivity() ndarray[tuple[int, ...], dtype[int64]][source]
Calculate how many times nodes appear in the edge adjacency map.
- Returns:
connectivity of each node
- Return type:
array[int] (num_nodes,)
- calculate_node_distances(repr='reduced') Tuple[ndarray, ndarray][source]
Calculate distances between all pairs of nodes.
- Parameters:
repr – ‘reduced’ or ‘transformed’
- Returns:
array of distances indices [num_pairs, 2]: indices of nodes corresponding to distances
- Return type:
distances [num_pairs]
- calculate_node_types() Dict[str, Set][source]
Calculate types of nodes from reduced representation
- Returns:
- a dictionary with sets of
corner_nodes (3 d.o.f. lie on UC boundary)
edge_nodes (2 d.o.f. lie on UC boundary)
face_nodes (1 d.o.f. lies on UC boundary)
inner_nodes (no d.o.f lie on UC boundary)
- Return type:
Dict[str, Set]
- calculate_relative_density(coords: str = 'transformed') float[source]
Calculate relative density of the lattice.
- Parameters:
coords (str, optional) – Use ‘transformed’ or ‘reduced’ coordinates. Defaults to ‘transformed’.
- Returns:
relative density of the lattice
- Return type:
float
- check_window_conditions() bool[source]
Operate on reduced edge adjacency representation.
- Check that
minimum reduced node coordinate is 0 and maximum is UC_L (1)
the only node types are face nodes and inner nodes
the connectivity of these nodes is 1
periodic partners come in pairs
See also
calculate_node_types(),calculate_nodal_connectivity(),check_periodic_partners()
- collapse_nodes_onto_boundaries(tol=0.0001)[source]
Collapse nodes which are close to boundaries onto the boundaries.
Operates on reduced nodal coordinates. All nodal coordinates which are very close to 0 or UC_L (within tolerance) will be replaced by 0 or UC_L, respectively.
- create_tesselated(nx: int, ny: int, nz: int, merge_colinear=True) Lattice[source]
Create periodic tesselation of the lattice
- Parameters:
nx (int) – number of times to repeat in x-direction
ny (int) – number of times to repeat in y-direction
nz (int) – number of times to repeat in z-direction
- Returns:
tesselated lattice
- Return type:
Examples
>>> nodes = [[0.2,0.3,0],[0.2,0,0],[1,0.3,0],[0.2,1,0],[0,0.3,0]] >>> edges = [[0,1],[0,2],[0,3],[0,4]] >>> lat = Lattice(nodal_positions=nodes, edge_adjacency=edges) >>> lt = lat.create_tesselated(2,2,1)
(
Source code,png,hires.png,pdf)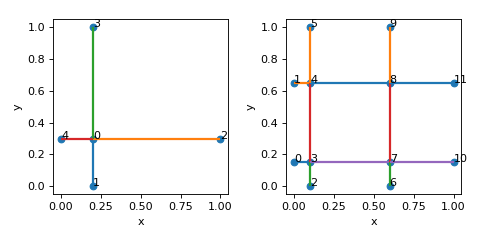
- create_windowed(max_num_attempts: int = 10, min_edge_length: float = 0.005, return_attempts: bool = False) Lattice[source]
Create a windowed representation of a lattice
- Returns:
a new lattice instance
- Return type:
Examples
>>> nodes = [[0,0,0],[1,0,0],[1,1,0],[0,1,0]] >>> edges = [[0,1],[1,2],[2,3],[3,0]] >>> lat = Lattice(nodal_positions=nodes, edge_adjacency=edges) >>> lat {'num_nodes': 4, 'num_edges': 4} >>> lat_w = lat.create_windowed() >>> lat_w {'num_nodes': 5, 'num_edges': 4}
(
Source code,png,hires.png,pdf)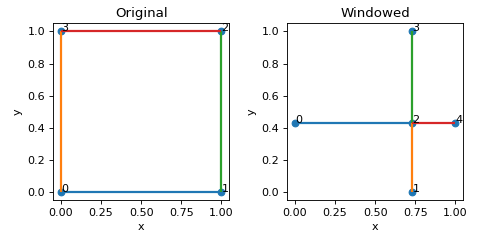
- crop_unit_cell(reduced_edge_coords: ndarray[tuple[int, ...], dtype[_ScalarType_co]] | None = None) None[source]
Crop lattice to fit within unit cell window.
Examples
>>> nodes = [[0,0,0],[1.5,0.5,0]] >>> edges = [[0,1]] >>> lat = Lattice(nodal_positions=nodes, edge_adjacency=edges) >>> lat.crop_unit_cell()
(
Source code,png,hires.png,pdf)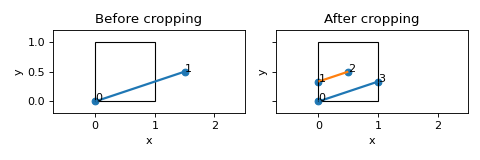
See also
- find_edge_intersections() List[Tuple[Set, ndarray[tuple[int, ...], dtype[_ScalarType_co]]]][source]
Find intersections between edge pairs.
Operates on reduced adjacency representation.
- Returns:
- dictionary with edge indices
as keys and coordinates of intersection points as values
- Return type:
edge_intersection_points
Example
>>> nodes = [[0,0,0],[1,0,0],[1,1,0],[0,1,0]] >>> edges = [[0,2],[1,3]] >>> lat = Lattice(nodal_positions=nodes, edge_adjacency=edges) >>> lat {'num_nodes': 4, 'num_edges': 2} >>> edge_intersections = lat.find_edge_intersections() >>> print(edge_intersections) [({0, 2}, array([[0.5, 0.5, 0. ]])), ({1, 3}, array([[0.5, 0.5, 0. ]]))]
(
Source code,png,hires.png,pdf)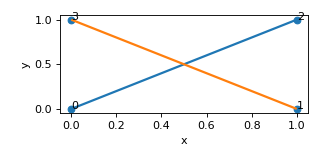
- find_nodes_on_edges() List[Tuple[Set, ndarray[tuple[int, ...], dtype[_ScalarType_co]]]][source]
Find nodes which lie on edges and are not endpoints.
Returns a list of tuples (edge: set, nodes: coordinates of points that need to split edges)
Example
>>> nodes = [[0,0,0],[1,0,0],[0.5,0,0]] >>> edges = [[0,1]] >>> lat = Lattice(nodal_positions=nodes, edge_adjacency=edges) >>> lat {'num_nodes': 3, 'num_edges': 1} >>> nodes_on_edges = lat.find_nodes_on_edges() >>> print(nodes_on_edges) [({0, 1}, array([[0.5, 0. , 0. ]]))]
(
Source code,png,hires.png,pdf)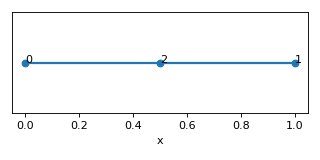
- get_periodic_partners() List[Set][source]
Calculate periodic partners.
Check is done first whether lattice is in a valid window condition.
- Returns:
list of 2-element sets of node numbers of periodic partners
- Return type:
list
Examples
Nodes {0,1} and {2,3} in the lattice below are periodic partners.
>>> nodes = [[0,0.5,0.5],[1,0.5,0.5],[0.5,0,0.5],[0.5,1,0.5],[0.5,0.5,0.5]] >>> edges = [[0,4],[1,4],[2,4],[3,4]] >>> lat = Lattice(nodal_positions=nodes, edge_adjacency=edges) >>> lat.get_periodic_partners() [{0, 1}, {2, 3}]
(
Source code,png,hires.png,pdf)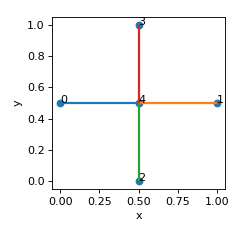
See also
- merge_colinear_edges() None[source]
Merge colinear edges based on nodal connectivity 2 and similar angle between two unit vectors along the edges.
Example
>>> nodes = [[0,0,0],[1,0,0],[0.5,0,0]] >>> edges = [[0,2],[1,2]] >>> lat = Lattice(nodal_positions=nodes, edge_adjacency=edges) >>> lat {'num_nodes': 3, 'num_edges': 2} >>> lat.merge_colinear_edges() >>> lat {'num_nodes': 2, 'num_edges': 1}
(
Source code,png,hires.png,pdf)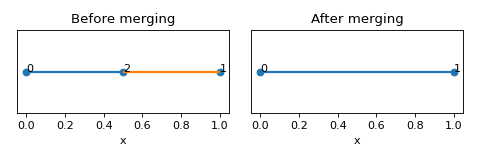
- merge_nonunique_nodes(decimals: int | None = None) None[source]
Merge nodes with identical coordinates and collapse self-incident edges
- obtain_shift_vector(max_num_attempts: int = 10, min_edge_length: float = 0.005, return_attempts: bool = False) ndarray[tuple[int, ...], dtype[_ScalarType_co]][source]
Get a shift vector for windowed representation.
- Raises:
WindowingException – _description_
WindowingException – _description_
- Returns:
_description_
- Return type:
npt.NDArray (3,)
- refine_mesh(min_length: float, min_div: int) Lattice[source]
Split edges into at least ‘min_div’ segments per edge and with each segment having length at least ‘min_length’
- remove_duplicate_edges_adjacency() None[source]
Remove duplicate edges.
Operates on the adjacency representation. Sort edges first
- remove_duplicate_edges_nodes() None[source]
Remove edges which are on top of each other.
Operates on edge coordinate representation. To deal with machine precision, round all nodal coordinates to a specific number of decimal places.
- remove_self_incident_edges() None[source]
Remove edges which have identical endpoints
Operates on the adjacency representation.
- set_fundamental_edge_radii(edge_radii: Iterable | float) None[source]
Set radii of fundamental edges.
- Parameters:
edge_radii (Union[Iterable, float]) – list of edge radii or single float
- split_edges_by_points(edge_split_coords: List[Tuple[Set, ndarray[tuple[int, ...], dtype[_ScalarType_co]]]]) None[source]
Split edges at specific coordinate points.
- Parameters:
edge_split_coords (List[Tuple[Set, npt.NDArray]]) – list of tuples (edge, point_array) in format as returned by functions
find_nodes_on_edges()andfind_edge_intersections(). Each edge will be split into multiple edges by the points in point_array.
Examples
>>> nodes = [[0,0,0],[1,0,0],[0.5,0,0]] >>> edges = [[0,1]] >>> lat = Lattice(nodal_positions=nodes, edge_adjacency=edges) >>> lat {'num_nodes': 3, 'num_edges': 1} >>> nodes_on_edges = lat.find_nodes_on_edges() >>> lat.split_edges_by_points(nodes_on_edges) >>> lat {'num_nodes': 3, 'num_edges': 2}
(
Source code,png,hires.png,pdf)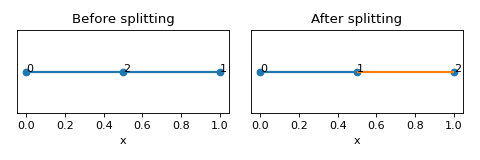
>>> nodes = [[0,0,0],[1,0,0],[1,1,0],[0,1,0]] >>> edges = [[0,2],[1,3]] >>> lat = Lattice(nodal_positions=nodes, edge_adjacency=edges) >>> lat {'num_nodes': 4, 'num_edges': 2} >>> edge_intersections = lat.find_edge_intersections() >>> lat.split_edges_by_points(edge_intersections) >>> lat {'num_nodes': 4, 'num_edges': 4}
(
Source code,png,hires.png,pdf)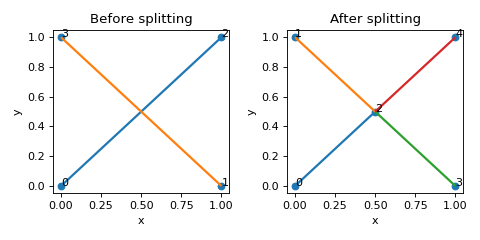
- to_dict(fundamental: bool = True) dict[source]
Obtain a dictionary with the reduced representation.
- transform_coordinates(coordinates: Iterable) ndarray[source]
Transform coordinates based on lattice constants.
Coordinates can be any O(3) vectors (e.g. node positions or edge vectors).
- Parameters:
coordinates (Iterable) – broadcastable into shape (N,3)
- Returns:
transformed coordinates
- Return type:
np.ndarray (N,3)
- property transform_matrix: ndarray[tuple[int, ...], dtype[float64]]
Assemble transformation matrix from crystal data.
Formula is in the Appendix to the PNAS paper: Lumpe, T. S. and Stankovic, T. (2020) https://www.pnas.org/doi/10.1073/pnas.2003504118.
- update_representations(edge_coords: ndarray[tuple[int, ...], dtype[_ScalarType_co]] | None = None) None[source]
Propagate one format of representation to all other available formats.
Implemented options for basis: - ‘reduced_edge_coords’
# TODO: update all relevant attributes
- property windowed_edge_radii: ndarray[tuple[int, ...], dtype[_ScalarType_co]]
Calculate edge radii for windowed edges from fundamental edge radii
- Returns:
edge radii for windowed edges
- Return type:
npt.NDArray
The Catalogue class
- class data.Catalogue(data: dict, indexing: int, **attrs)[source]
Bases:
objectUnit cell catalogue object.
Note
- Two class methods are available to initialise the object:
from_file()Used to read the catalogue from a file
>>> cat = Catalogue.from_file('Unit_Cell_Catalog.txt', 1)
from_dict()Used to create the catalogue from unit cells either from scratch or when unit cells from a file are modified
>>> nodes = [[0,0,0],[1,0,0],[0.5,1,0],[0.5,1,1]] >>> edges = [[0,1],[1,2],[0,2],[0,3],[1,3],[2,3]] >>> lat = Lattice(nodal_positions=nodes, edge_adjacency=edges) >>> lat {'num_nodes': 4, 'num_edges': 6} >>> cat_dict = {'pyramid':lat.to_dict()} >>> cat = Catalogue.from_dict(cat_dict)
- See Also:
Lattice.to_dict()
- classmethod from_dict(lattice_dicts: Dict[str, Dict]) Catalogue[source]
Generate unit cell catalogue from dictionary representation
- Parameters:
lattice_dicts (Dict[str, Dict]) – dictionary of lattice dictionaries
- Returns:
Catalogue
Note
- Lattice dictionaries must contain the following keys:
edge_adjacency
nodal_positions or reduced_node_coordinates
- They can contain also:
lattice_constants
compliance_tensors # assuming Voigt notation, legacy support
compliance_tensors_V # Voigt notation
compliance_tensors_M # Mandel notation
fundamental_edge_adjacency
fundamental_tesselation_vecs
fundamental_edge_radii
base_name
imperfection_kind
imperfection_level
nodal_hash
- classmethod from_file(fn: str, indexing: int, progress: bool = False, regex: str = None) Catalogue[source]
Read catalogue from a file.
- Parameters:
fn (str) – path to input file
indexing (int) – 0 or 1 as the basis of edge indexing
progress (bool, optional) – whether to show progress bar. Defaults to False.
regex (str, optional) – regular expression to filter names. Defaults to None.
- Returns:
Catalogue
- classmethod get_names(fn: str) List[str][source]
Retrieve names from catalogue without reading it into memory.
- Parameters:
fn (str) – path to input file
- Returns:
list of names
- Return type:
List[str]
- get_unit_cell(name: str) dict[source]
Return a dictionary which represents unit cell.
- Parameters:
name (str) – Name of the unit cell from the catalogue that will be returned
- Returns:
Dictionary describing the unit cell
- Return type:
dict
Note
- Returned dictionary contains all available keys from the following:
name
lattice constants: [a,b,c,alpha,beta,gamma]
average connectivity
compliance_tensors_M: dictionary {rel_dens: compliance tensor}
compliance_tensors_V: dictionary {rel_dens: compliance tensor}
nodal_positions: nested list of shape (num_nodes, 3)
edge_adjacency: nested list of shape (num_edges, 2) (0-indexed)
fundamental_edge_adjacency: nested list of shape (num_fundamental_edges, 2) (0-indexed)
fundamental_tesselation_vecs: nested list of shape (num_fundamental_edges, 6) or (num_fundamental_edges, 3) (0-indexed)
fundamental_edge_radii: dictionary {rel_dens: list of edge radii}
The dictionary can be unpacked in the creation of a Lattice object
>>> from data import Lattice, Catalogue >>> cat = Catalogue.from_file('Unit_Cell_Catalog.txt', 1) >>> lat = Lattice(**cat.get_unit_cell(cat.names[0])) >>> lat {'name': 'cub_Z06.0_E1', 'num_nodes': 8, 'num_edges': 12}
See also
data.Lattice.__init__()
- static n_2_bn(name: str) str[source]
Convert name of a unit cell to base name.
- Relies on the following naming convention:
[base_name]_p_[imperfection_level]_[nodal_hash]
- where base_name is of the form:
[symmetry]_[connectivity]_[code]
- For instance:
cub_Z12.0_E19_p_0.0_9113732828474860344
- Parameters:
name (str) – Name of the unit cell
- Returns:
Base name of the unit cell
- Return type:
str